Published in Nature, November 5, 2025
A cell census of the developing human and mammalian brain
How does the brain build itself? What sparks the transformation from a handful of cells into a complex organ that powers thought, emotion, and behavior?
This ground-breaking publication from the BRAIN Initiative Cell Census Network (BICCN) and BRAIN Initiative Cell Atlas Network (BICAN) brings us closer to answering those questions. It’s the result of a massive scientific collaboration, spanning multiple institutions and disciplines, mapping brain development in extraordinary detail.
Webinar: Hear from the authors!



About
-
A collection of papers in Nature journals and 2 companion papers
-
Supported by the National Institutes of Health BRAIN Initiative with funding from BICCN and BICAN awards.
-
Includes spatial and single cell omics datasets from multiple species, a 3D developmental mouse Common Coordinate Framework, and perturbation and lineage tracing.
Scientific Publications
Nowakowski et al., Nature
The new frontier in understanding human and mammalian brain development
Nowakowski, T., et al. The new frontier of human and mammalian brain development. Nature (2025). https://doi.org/10.1038/s41586-025-09652-1
perspective
Wang et al., Nature
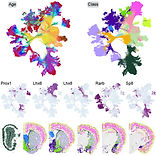
Molecular and cellular dynamics of the developing human neocortex
Wang, L., Wang, C., Moriano, J.A. et al. Molecular and cellular dynamics of the developing human neocortex. Nature (2025). https://doi.org/10.1038/s41586-024-08351-7
core
Keefe et al., Nature
Lineage-resolved atlas of the developing human cortex
Keefe, M.G., Steyert, M.R., et al. Developmental lineage relationships of radial glial cells. Nature (2025) https://doi.org/10.1038/s41586-025-09033-8
core
Kaplan et al., Nature
Sensory input, sex and function shape hypothalamic cell type development
Kaplan, H.S., Logeman, B.L., Zhang, K. et al. Sensory input, sex and function shape hypothalamic cell type development. Nature (2025). https://doi.org/10.1038/s41586-025-08603-0
core
Gao et al., Nature
Continuous cell-type diversification in mouse visual cortex development
Gao, Y., et al. Continuous cell-type diversification in mouse visual cortex development. Nature (2025). https://doi.org/10.1038/s41586-025-09644-1
core
Corrigan et al., Nature
Conservation and alteration of mammalian striatal interneurons
Corrigan, E., et al. Conservation and alteration of mammalian striatal interneurons. Nature (2025). https://doi.org/10.1038/s41586-025-09592-w
core
Mannens et al., Nature
Chromatin accessibility during human first-trimester neurodevelopment
Mannens, C.A., et al. Chromatin accessibility during human first-trimester neurodevelopment. Nature (2024). https://doi.org/10.1038/s41586-024-07234-1
core
Zhang et al., Nature
Spatial dynamics of brain development and neuroinflammation
Zhang, D., et al. Spatial dynamics of brain development and neuroinflammation. Nature (2025). https://doi.org/10.1038/s41586-025-09663-y
core
van Velthoven et al., Nature
Transcriptomic and spatial organization of telencephalic GABAergic neurons
van Velthoven, C.T.J., et al. The transcriptomic and spatial organization of telencephalic GABAergic neuronal types. Nature (2025). https://doi.org/10.1038/s41586-025-09296-1
core
Kronman et al., Nature Communications
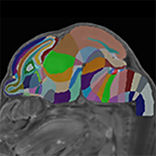
Developmental mouse brain common coordinate framework
Kronman, F.N., Liwang, J.K., Betty, R. et al. Developmental mouse brain common coordinate framework. Nat Commun 15, 9072 (2024). https://doi.org/10.1038/s41467-024-53254-w
core
Nano et al., Nature Neuroscience
Integrated analysis of molecular atlases unveils modules driving developmental cell subtype specification in the human cortex
Nano, P.R., Fazzari, E., Azizad, D. et al. Integrated analysis of molecular atlases unveils modules driving developmental cell subtype specification in the human cortex. Nat Neurosci 28, 949–963 (2025). https://doi.org/10.1038/s41593-025-01933-2
core
Chen et al., Nature
Whole-cortex in situ sequencing reveals input-dependent area identity
Chen, X., Fischer, S., Rue, M.C.P. et al. Whole-cortex in situ sequencing reveals input-dependent area identity. Nature (2024). https://doi.org/10.1038/s41586-024-07221-6
core
Werner et al., PLoS Biology
Meta-analysis of single-cell RNA sequencing co-expression in human neural organoids reveals their high variability in recapitulating primary tissue
Werner, J.M. and Gillis, J. Meta-analysis of single-cell RNA sequencing co-expression in human neural organoids reveals their high variability in recapitulating primary tissue. PLoS Biology (2024). https://doi.org/10.1371/journal.pbio.3002912